Phytoplankton nutrient limitation maps
Contents
Phytoplankton nutrient limitation maps#
%load_ext autoreload
%autoreload 2
import xarray as xr
from dask.distributed import Client
import catalog
import util
xr.set_options(keep_attrs=True)
import matplotlib.pyplot as plt
import cartopy
import cartopy.crs as ccrs
import pop_tools
import utils
import numpy as np
import pandas as pd
/glade/work/kristenk/miniconda/envs/cesm-exp/lib/python3.7/site-packages/dask_jobqueue/core.py:20: FutureWarning: tmpfile is deprecated and will be removed in a future release. Please use dask.utils.tmpfile instead.
from distributed.utils import tmpfile
ds_grid = pop_tools.get_grid('POP_gx1v7')
lons=ds_grid.TLONG
lats=ds_grid.TLAT
Parameters#
casename = 'g.e22.GOMIPECOIAF_JRA-1p4-2018.TL319_g17.4p2z.001'
component = 'pop'
stream = 'h'
cluster_scheduler_address = None
# Parameters
casename = "g.e22.GOMIPECOIAF_JRA-1p4-2018.TL319_g17.4p2z.001branch"
component = "pop"
stream = "h"
cluster_scheduler_address = "tcp://10.12.206.3:35506"
assert component in ['pop']
assert stream in ['h', 'h.ecosys.nday1']
Connect to cluster#
if cluster_scheduler_address is None:
cluster, client = util.get_ClusterClient()
cluster.scale(12)
else:
client = Client(cluster_scheduler_address)
client
Client
Client-efd765cc-9357-11ed-a020-3cecef1acc46
| Connection method: Direct | |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/8787/status |
Scheduler Info
Scheduler
Scheduler-7b28f886-17c1-40f8-ae24-3360603786e8
| Comm: tcp://10.12.206.3:35506 | Workers: 32 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/8787/status | Total threads: 32 |
| Started: 21 minutes ago | Total memory: 744.96 GiB |
Workers
Worker: PBSCluster-0
| Comm: tcp://10.12.206.60:40381 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/37473/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.60:43642 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-she593m8 | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 0.0% | Last seen: Just now |
| Memory usage: 459.22 MiB | Spilled bytes: 0 B |
| Read bytes: 29.97 kiB | Write bytes: 20.14 kiB |
Worker: PBSCluster-1
| Comm: tcp://10.12.206.57:34913 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/43168/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.57:40520 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-ais4w5nl | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 515.51 MiB | Spilled bytes: 0 B |
| Read bytes: 37.49 kiB | Write bytes: 8.19 MiB |
Worker: PBSCluster-10
| Comm: tcp://10.12.206.55:40610 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/41580/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.55:41770 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-24q12p4a | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 509.34 MiB | Spilled bytes: 0 B |
| Read bytes: 23.58 kiB | Write bytes: 25.50 kiB |
Worker: PBSCluster-11
| Comm: tcp://10.12.206.35:36760 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/34231/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.35:39163 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-tm589a2g | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 570.82 MiB | Spilled bytes: 0 B |
| Read bytes: 25.08 kiB | Write bytes: 12.32 kiB |
Worker: PBSCluster-12
| Comm: tcp://10.12.206.42:46667 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/44609/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.42:43503 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-63bl396p | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 0.0% | Last seen: Just now |
| Memory usage: 483.36 MiB | Spilled bytes: 0 B |
| Read bytes: 29.04 kiB | Write bytes: 11.90 kiB |
Worker: PBSCluster-13
| Comm: tcp://10.12.206.55:34741 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/41987/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.55:39933 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-3taktpky | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 485.52 MiB | Spilled bytes: 0 B |
| Read bytes: 22.73 kiB | Write bytes: 21.44 kiB |
Worker: PBSCluster-14
| Comm: tcp://10.12.206.42:44723 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/38884/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.42:37139 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-btgwv71o | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 534.16 MiB | Spilled bytes: 0 B |
| Read bytes: 28.91 kiB | Write bytes: 15.02 kiB |
Worker: PBSCluster-15
| Comm: tcp://10.12.206.60:34840 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/36356/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.60:36517 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-s50upym9 | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 4.0% | Last seen: Just now |
| Memory usage: 481.70 MiB | Spilled bytes: 0 B |
| Read bytes: 19.66 kiB | Write bytes: 10.14 kiB |
Worker: PBSCluster-16
| Comm: tcp://10.12.206.55:33480 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/44604/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.55:34515 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-20gqvxq8 | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 493.89 MiB | Spilled bytes: 0 B |
| Read bytes: 23.65 kiB | Write bytes: 23.37 kiB |
Worker: PBSCluster-17
| Comm: tcp://10.12.206.42:46005 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/44117/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.42:32947 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-18iilri4 | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 0.0% | Last seen: Just now |
| Memory usage: 521.43 MiB | Spilled bytes: 0 B |
| Read bytes: 29.05 kiB | Write bytes: 13.52 kiB |
Worker: PBSCluster-18
| Comm: tcp://10.12.206.60:33226 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/42773/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.60:46337 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-8qq6z3y3 | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 484.48 MiB | Spilled bytes: 0 B |
| Read bytes: 19.67 kiB | Write bytes: 10.15 kiB |
Worker: PBSCluster-19
| Comm: tcp://10.12.206.36:36934 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/38605/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.36:40345 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-0wibvgh_ | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 0.0% | Last seen: Just now |
| Memory usage: 522.61 MiB | Spilled bytes: 0 B |
| Read bytes: 126.49 kiB | Write bytes: 96.73 kiB |
Worker: PBSCluster-2
| Comm: tcp://10.12.206.42:35532 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/37233/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.42:38029 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-tvn8qi89 | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 471.90 MiB | Spilled bytes: 0 B |
| Read bytes: 29.04 kiB | Write bytes: 13.51 kiB |
Worker: PBSCluster-20
| Comm: tcp://10.12.206.57:44618 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/35828/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.57:39590 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-j87b3i6z | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 0.0% | Last seen: Just now |
| Memory usage: 489.03 MiB | Spilled bytes: 0 B |
| Read bytes: 37.55 kiB | Write bytes: 8.20 MiB |
Worker: PBSCluster-21
| Comm: tcp://10.12.206.42:36146 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/42013/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.42:42567 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-te65hpsa | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 544.07 MiB | Spilled bytes: 0 B |
| Read bytes: 28.85 kiB | Write bytes: 17.41 kiB |
Worker: PBSCluster-22
| Comm: tcp://10.12.206.37:45266 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/41617/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.37:46778 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-6zjl_rf2 | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 496.08 MiB | Spilled bytes: 0 B |
| Read bytes: 188.98 MiB | Write bytes: 200.39 kiB |
Worker: PBSCluster-23
| Comm: tcp://10.12.206.57:36646 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/33282/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.57:39220 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-tedt0oqb | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 0.0% | Last seen: Just now |
| Memory usage: 524.25 MiB | Spilled bytes: 0 B |
| Read bytes: 37.47 kiB | Write bytes: 8.19 MiB |
Worker: PBSCluster-24
| Comm: tcp://10.12.206.35:43781 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/41280/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.35:41682 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-oenmf7ye | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 470.49 MiB | Spilled bytes: 0 B |
| Read bytes: 24.97 kiB | Write bytes: 13.94 kiB |
Worker: PBSCluster-25
| Comm: tcp://10.12.206.42:38958 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/45762/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.42:41096 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-r999w54s | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 461.88 MiB | Spilled bytes: 0 B |
| Read bytes: 28.91 kiB | Write bytes: 14.21 kiB |
Worker: PBSCluster-26
| Comm: tcp://10.12.206.35:41019 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/46197/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.35:41093 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-n8z_ugpj | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 542.76 MiB | Spilled bytes: 0 B |
| Read bytes: 25.01 kiB | Write bytes: 13.96 kiB |
Worker: PBSCluster-27
| Comm: tcp://10.12.206.40:37756 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/34868/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.40:46061 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-ah4h4uwy | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 492.45 MiB | Spilled bytes: 0 B |
| Read bytes: 24.94 kiB | Write bytes: 14.52 kiB |
Worker: PBSCluster-28
| Comm: tcp://10.12.206.55:40536 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/40763/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.55:43688 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-mqh1_swd | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 478.43 MiB | Spilled bytes: 0 B |
| Read bytes: 22.70 kiB | Write bytes: 21.42 kiB |
Worker: PBSCluster-29
| Comm: tcp://10.12.206.35:43010 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/45869/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.35:38155 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-0zp2yifw | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 514.69 MiB | Spilled bytes: 0 B |
| Read bytes: 25.33 kiB | Write bytes: 11.52 kiB |
Worker: PBSCluster-3
| Comm: tcp://10.12.206.55:43243 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/45154/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.55:40506 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-d77lpe1m | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 0.0% | Last seen: Just now |
| Memory usage: 474.66 MiB | Spilled bytes: 0 B |
| Read bytes: 22.71 kiB | Write bytes: 19.81 kiB |
Worker: PBSCluster-30
| Comm: tcp://10.12.206.42:40059 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/38562/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.42:35150 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-s1j98ibm | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 4.0% | Last seen: Just now |
| Memory usage: 493.88 MiB | Spilled bytes: 0 B |
| Read bytes: 29.18 kiB | Write bytes: 12.00 kiB |
Worker: PBSCluster-31
| Comm: tcp://10.12.206.55:40509 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/46123/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.55:42505 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-0193oowl | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 451.69 MiB | Spilled bytes: 0 B |
| Read bytes: 23.02 kiB | Write bytes: 23.14 kiB |
Worker: PBSCluster-4
| Comm: tcp://10.12.206.60:33761 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/38547/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.60:43086 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-go_20xsh | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 509.03 MiB | Spilled bytes: 0 B |
| Read bytes: 29.96 kiB | Write bytes: 20.13 kiB |
Worker: PBSCluster-5
| Comm: tcp://10.12.206.56:36183 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/39960/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.56:36523 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-ak_et0qx | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 490.81 MiB | Spilled bytes: 0 B |
| Read bytes: 387.03 MiB | Write bytes: 199.60 kiB |
Worker: PBSCluster-6
| Comm: tcp://10.12.206.55:42418 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/43316/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.55:35755 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-w069mt42 | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 461.57 MiB | Spilled bytes: 0 B |
| Read bytes: 24.01 kiB | Write bytes: 23.52 kiB |
Worker: PBSCluster-7
| Comm: tcp://10.12.206.42:38064 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/33959/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.42:37237 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-8g92ok8a | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 476.61 MiB | Spilled bytes: 0 B |
| Read bytes: 28.93 kiB | Write bytes: 15.84 kiB |
Worker: PBSCluster-8
| Comm: tcp://10.12.206.35:46716 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/38950/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.35:46823 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-t0bj5mka | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 494.70 MiB | Spilled bytes: 0 B |
| Read bytes: 25.02 kiB | Write bytes: 12.35 kiB |
Worker: PBSCluster-9
| Comm: tcp://10.12.206.55:36074 | Total threads: 1 |
| Dashboard: https://jupyterhub.hpc.ucar.edu/stable/user/kristenk/proxy/41381/status | Memory: 23.28 GiB |
| Nanny: tcp://10.12.206.55:35112 | |
| Local directory: /glade/scratch/kristenk/dask-workers/dask-worker-space/worker-vzrt3wyj | |
| Tasks executing: 0 | Tasks in memory: 0 |
| Tasks ready: 0 | Tasks in flight: 0 |
| CPU usage: 2.0% | Last seen: Just now |
| Memory usage: 601.68 MiB | Spilled bytes: 0 B |
| Read bytes: 22.70 kiB | Write bytes: 19.80 kiB |
Load the data#
dsets = catalog.to_dataset_dict(
case=casename,
component=component,
stream=stream,
)
dsets.keys()
g.e22.GOMIPECOIAF_JRA-1p4-2018.TL319_g17.4p2z.001branch pop h /glade/scratch/kristenk/archive
dict_keys(['g.e22.GOMIPECOIAF_JRA-1p4-2018.TL319_g17.4p2z.001branch.pop.h'])
Compute long-term mean and plot#
ds = dsets[f'{casename}.{component}.{stream}']
Variable list#
variables =['sp_Fe_lim_Cweight_avg_100m','sp_P_lim_Cweight_avg_100m','sp_N_lim_Cweight_avg_100m',
'diat_Fe_lim_Cweight_avg_100m', 'diat_P_lim_Cweight_avg_100m','diat_N_lim_Cweight_avg_100m',
'diat_SiO3_lim_Cweight_avg_100m','diaz_P_lim_Cweight_avg_100m',
'diaz_Fe_lim_Cweight_avg_100m','cocco_Fe_lim_Cweight_avg_100m','cocco_C_lim_Cweight_avg_100m','cocco_N_lim_Cweight_avg_100m',
'cocco_P_lim_Cweight_avg_100m','sp_Fe_lim_surf','sp_P_lim_surf','sp_N_lim_surf',
'diat_Fe_lim_surf', 'diat_P_lim_surf','diat_N_lim_surf','diat_SiO3_lim_surf',
'diaz_P_lim_surf','cocco_Fe_lim_surf','cocco_C_lim_surf','cocco_N_lim_surf',
'cocco_P_lim_surf','diaz_Fe_lim_surf']
keep_vars=['z_t','z_t_150m','dz','time_bound','time','TAREA','TLAT','TLONG'] + variables
ds = ds.drop([v for v in ds.variables if v not in keep_vars])
Make a monthly climatology dataset#
time = pd.date_range(start="2009-01-01", end="2019-01-01",freq="M")
mon_ds = ds.copy()
mon_ds['time'] = time
mon_ds = ds.groupby('time.month').mean('time')
Compute long-term mean#
dt_wgt = ds.time_bound.diff('d2').squeeze()
dt_wgt /= dt_wgt.sum()
dt_wgt
<xarray.DataArray 'time_bound' (time: 120)>
dask.array<truediv, shape=(120,), dtype=float64, chunksize=(1,), chunktype=numpy.ndarray>
Coordinates:
* time (time) object 2012-01-16 12:00:00 ... 2021-12-16 12:00:00
Attributes:
long_name: boundaries for time-averaging interval
units: days since 0000-01-01 00:00:00ds = ds.weighted(dt_wgt).mean('time')
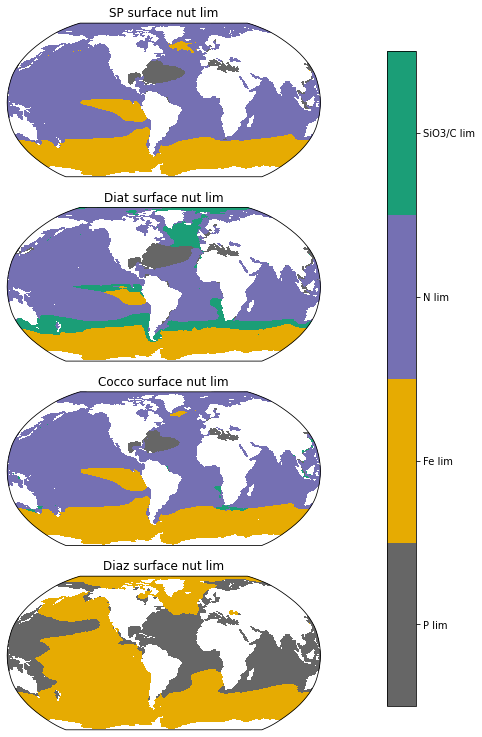
Phytoplankton limitations at the surface#
#most limiting nutrient - concatenate the limitation terms so that nutrient limitation terms are each along the 'nutrient' dimension
# 0 = PO4
# 1 = Fe
# 2 = NO3 (only for sp and diat)
# 3 = Si (only for diat)
# 3 = C (only for cocco)
limarray_sp=xr.concat((ds.sp_P_lim_surf, ds.sp_Fe_lim_surf,ds.sp_N_lim_surf),dim='nutrient')
limarray_diat=xr.concat((ds.diat_P_lim_surf, ds.diat_Fe_lim_surf, ds.diat_N_lim_surf, ds.diat_SiO3_lim_surf),dim='nutrient')
limarray_diaz=xr.concat((ds.diaz_P_lim_surf, ds.diaz_Fe_lim_surf),dim='nutrient')
limarray_cocco=xr.concat((ds.cocco_P_lim_surf, ds.cocco_Fe_lim_surf, ds.cocco_N_lim_surf, ds.cocco_C_lim_surf),dim='nutrient')
most_lim_sp=limarray_sp.argmin(dim='nutrient', skipna=False).squeeze()
most_lim_diat=limarray_diat.argmin(dim='nutrient', skipna=False).squeeze()
most_lim_diaz=limarray_diaz.argmin(dim='nutrient', skipna=False).squeeze()
most_lim_cocco=limarray_cocco.argmin(dim='nutrient', skipna=False).squeeze()
mask = np.isnan(ds.sp_N_lim_surf.squeeze())
fig = plt.figure(figsize=(8,13))
colorbar_specs = {'ticks' : np.arange(0,4,1)}
ax = fig.add_subplot(4,1,2, projection=ccrs.Robinson(central_longitude=305.0))
ax.set_title('Diat surface nut lim', fontsize=12)
lon, lat, field = utils.adjust_pop_grid(lons, lats, most_lim_diat.where(~mask))
pc=ax.pcolormesh(lon, lat, field, cmap=plt.cm.get_cmap('Dark2_r',4),vmin=-0.5,vmax=3.5,transform=ccrs.PlateCarree())
ax = fig.add_subplot(4,1,1, projection=ccrs.Robinson(central_longitude=305.0))
ax.set_title('SP surface nut lim', fontsize=12)
lon, lat, field = utils.adjust_pop_grid(lons, lats, most_lim_sp.where(~mask))
pc=ax.pcolormesh(lon, lat, field, cmap=plt.cm.get_cmap('Dark2_r',4),vmin=-0.5,vmax=3.5,transform=ccrs.PlateCarree())
ax = fig.add_subplot(4,1,3, projection=ccrs.Robinson(central_longitude=305.0))
ax.set_title('Cocco surface nut lim', fontsize=12)
lon, lat, field = utils.adjust_pop_grid(lons, lats, most_lim_cocco.where(~mask))
pc=ax.pcolormesh(lon, lat, field, cmap=plt.cm.get_cmap('Dark2_r',4),vmin=-0.5,vmax=3.5,transform=ccrs.PlateCarree())
ax = fig.add_subplot(4,1,4, projection=ccrs.Robinson(central_longitude=305.0))
ax.set_title('Diaz surface nut lim', fontsize=12)
lon, lat, field = utils.adjust_pop_grid(lons, lats, most_lim_diaz.where(~mask))
pc=ax.pcolormesh(lon, lat, field, cmap=plt.cm.get_cmap('Dark2_r',4),vmin=-0.5,vmax=3.5,transform=ccrs.PlateCarree())
fig.subplots_adjust(right=0.8)
cbar_ax = fig.add_axes([0.85, 0.15, 0.05, 0.7])
cbar = fig.colorbar(pc, cax=cbar_ax,**colorbar_specs)
cbar.ax.set_yticklabels(['P lim', 'Fe lim', 'N lim','SiO3/C lim']);
Biomass weighted phytoplankton limitations in the top 100m#
#most limiting nutrient - concatenate the limitation terms so that nutrient limitation terms are each along the 'nutrient' dimension
# 0 = PO4
# 1 = Fe
# 2 = NO3 (only for sp and diat)
# 3 = Si (only for diat)
# 3 = C (only for cocco)
limarray_sp=xr.concat((ds.sp_P_lim_Cweight_avg_100m, ds.sp_Fe_lim_Cweight_avg_100m,ds.sp_N_lim_Cweight_avg_100m),dim='nutrient')
limarray_diat=xr.concat((ds.diat_P_lim_Cweight_avg_100m, ds.diat_Fe_lim_Cweight_avg_100m, ds.diat_N_lim_Cweight_avg_100m, ds.diat_SiO3_lim_Cweight_avg_100m),dim='nutrient')
limarray_diaz=xr.concat((ds.diaz_P_lim_Cweight_avg_100m, ds.diaz_Fe_lim_Cweight_avg_100m),dim='nutrient')
limarray_cocco=xr.concat((ds.cocco_P_lim_Cweight_avg_100m, ds.cocco_Fe_lim_Cweight_avg_100m, ds.cocco_N_lim_Cweight_avg_100m, ds.cocco_C_lim_Cweight_avg_100m),dim='nutrient')
most_lim_sp=limarray_sp.argmin(dim='nutrient', skipna=False).squeeze()
most_lim_diat=limarray_diat.argmin(dim='nutrient', skipna=False).squeeze()
most_lim_diaz=limarray_diaz.argmin(dim='nutrient', skipna=False).squeeze()
most_lim_cocco=limarray_cocco.argmin(dim='nutrient', skipna=False).squeeze()
mask = np.isnan(ds.sp_N_lim_surf.squeeze())
fig = plt.figure(figsize=(8,13))
colorbar_specs = {'ticks' : np.arange(0,4,1)}
ax = fig.add_subplot(4,1,2, projection=ccrs.Robinson(central_longitude=305.0))
ax.set_title('Diat biomass-weighted nut lim', fontsize=12)
lon, lat, field = utils.adjust_pop_grid(lons, lats, most_lim_diat.where(~mask))
pc=ax.pcolormesh(lon, lat, field, cmap=plt.cm.get_cmap('Dark2_r',4),vmin=-0.5,vmax=3.5,transform=ccrs.PlateCarree())
ax = fig.add_subplot(4,1,1, projection=ccrs.Robinson(central_longitude=305.0))
ax.set_title('SP biomass-weighted nut lim', fontsize=12)
lon, lat, field = utils.adjust_pop_grid(lons, lats, most_lim_sp.where(~mask))
pc=ax.pcolormesh(lon, lat, field, cmap=plt.cm.get_cmap('Dark2_r',4),vmin=-0.5,vmax=3.5,transform=ccrs.PlateCarree())
ax = fig.add_subplot(4,1,3, projection=ccrs.Robinson(central_longitude=305.0))
ax.set_title('Cocco biomass-weighted nut lim', fontsize=12)
lon, lat, field = utils.adjust_pop_grid(lons, lats, most_lim_cocco.where(~mask))
pc=ax.pcolormesh(lon, lat, field, cmap=plt.cm.get_cmap('Dark2_r',4),vmin=-0.5,vmax=3.5,transform=ccrs.PlateCarree())
ax = fig.add_subplot(4,1,4, projection=ccrs.Robinson(central_longitude=305.0))
ax.set_title('Diaz biomass-weighted nut lim', fontsize=12)
lon, lat, field = utils.adjust_pop_grid(lons, lats, most_lim_diaz.where(~mask))
pc=ax.pcolormesh(lon, lat, field, cmap=plt.cm.get_cmap('Dark2_r',4),vmin=-0.5,vmax=3.5,transform=ccrs.PlateCarree())
fig.subplots_adjust(right=0.8)
cbar_ax = fig.add_axes([0.85, 0.15, 0.05, 0.7])
cbar = fig.colorbar(pc, cax=cbar_ax,**colorbar_specs)
cbar.ax.set_yticklabels(['P lim', 'Fe lim', 'N lim','SiO3/C lim']);

Monthly climatology of surface phytoplankton limitations#
#most limiting nutrient - concatenate the limitation terms so that nutrient limitation terms are each along the 'nutrient' dimension
# 0 = PO4
# 1 = Fe
# 2 = NO3 (only for sp and diat)
# 3 = Si (only for diat)
# 3 = C (only for cocco)
limarray_sp=xr.concat((mon_ds.sp_P_lim_surf, mon_ds.sp_Fe_lim_surf,mon_ds.sp_N_lim_surf),dim='nutrient')
limarray_diat=xr.concat((mon_ds.diat_P_lim_surf, mon_ds.diat_Fe_lim_surf, mon_ds.diat_N_lim_surf, mon_ds.diat_SiO3_lim_surf),dim='nutrient')
limarray_diaz=xr.concat((mon_ds.diaz_P_lim_surf, mon_ds.diaz_Fe_lim_surf),dim='nutrient')
limarray_cocco=xr.concat((mon_ds.cocco_P_lim_surf, mon_ds.cocco_Fe_lim_surf, mon_ds.cocco_N_lim_surf, mon_ds.cocco_C_lim_surf),dim='nutrient')
most_lim_sp=limarray_sp.argmin(dim='nutrient', skipna=False)
most_lim_diat=limarray_diat.argmin(dim='nutrient', skipna=False)
most_lim_diaz=limarray_diaz.argmin(dim='nutrient', skipna=False)
most_lim_cocco=limarray_cocco.argmin(dim='nutrient', skipna=False)
mask = np.isnan(ds.sp_N_lim_surf.squeeze())
fig = plt.figure(figsize=(12,23))
for row in np.arange(1,13):
ts=row-1
plot = row*3 - 2
#row 1 Jan
ax = fig.add_subplot(12,3,plot, projection=ccrs.Robinson(central_longitude=305.0))
ax.set_title('Diat surface nut lim', fontsize=12)
lon, lat, field = utils.adjust_pop_grid(lons, lats, most_lim_diat.isel(month=ts).where(~mask))
pc=ax.pcolormesh(lon, lat, field, cmap=plt.cm.get_cmap('Dark2_r',4),vmin=-0.5,vmax=3.5,transform=ccrs.PlateCarree())
colorbar_specs = {'ticks' : np.arange(0,4,1)}
plot = row*3 - 1
ax = fig.add_subplot(12,3,plot, projection=ccrs.Robinson(central_longitude=305.0))
ax.set_title('SP surface nut lim', fontsize=12)
lon, lat, field = utils.adjust_pop_grid(lons, lats, most_lim_sp.isel(month=ts).where(~mask))
pc=ax.pcolormesh(lon, lat, field, cmap=plt.cm.get_cmap('Dark2_r',4),vmin=-0.5,vmax=3.5,transform=ccrs.PlateCarree())
colorbar_specs = {'ticks' : np.arange(0,4,1)}
plot = row*3
ax = fig.add_subplot(12,3,plot, projection=ccrs.Robinson(central_longitude=305.0))
ax.set_title('Cocco surface nut lim', fontsize=12)
lon, lat, field = utils.adjust_pop_grid(lons, lats, most_lim_cocco.isel(month=ts).where(~mask))
pc=ax.pcolormesh(lon, lat, field, cmap=plt.cm.get_cmap('Dark2_r',4),vmin=-0.5,vmax=3.5,transform=ccrs.PlateCarree())
colorbar_specs = {'ticks' : np.arange(0,4,1)}
fig.subplots_adjust(right=0.8)
cbar_ax = fig.add_axes([0.85, 0.15, 0.05, 0.7])
cbar = fig.colorbar(pc, cax=cbar_ax,**colorbar_specs)
cbar.ax.set_yticklabels(['P lim', 'Fe lim', 'N lim','SiO3/C lim']);

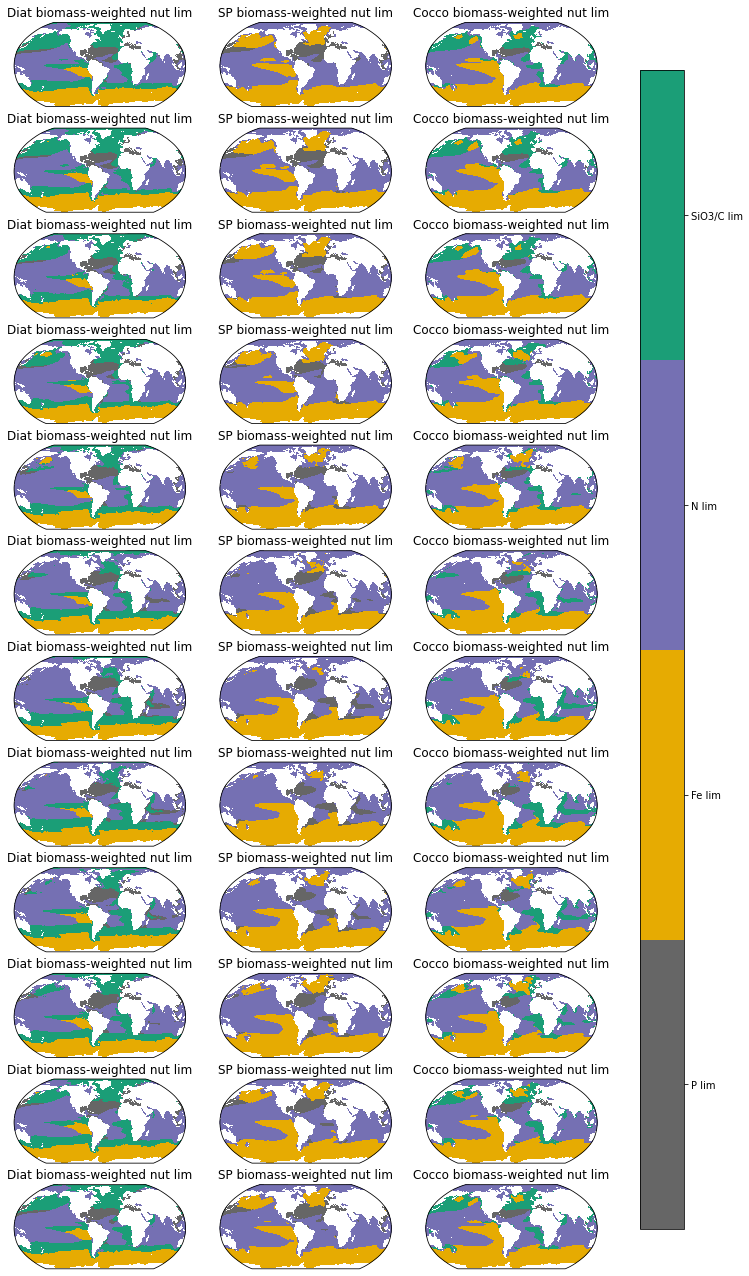
Monthly climatology of biomass-weighted phytoplankton nutrient limitation terms over the top 100m#
#most limiting nutrient - concatenate the limitation terms so that nutrient limitation terms are each along the 'nutrient' dimension
# 0 = PO4
# 1 = Fe
# 2 = NO3 (only for sp and diat)
# 3 = Si (only for diat)
# 3 = C (only for cocco)
limarray_sp=xr.concat((mon_ds.sp_P_lim_Cweight_avg_100m, mon_ds.sp_Fe_lim_Cweight_avg_100m,mon_ds.sp_N_lim_Cweight_avg_100m),dim='nutrient')
limarray_diat=xr.concat((mon_ds.diat_P_lim_Cweight_avg_100m, mon_ds.diat_Fe_lim_Cweight_avg_100m, mon_ds.diat_N_lim_Cweight_avg_100m, mon_ds.diat_SiO3_lim_Cweight_avg_100m),dim='nutrient')
limarray_diaz=xr.concat((mon_ds.diaz_P_lim_Cweight_avg_100m, mon_ds.diaz_Fe_lim_Cweight_avg_100m),dim='nutrient')
limarray_cocco=xr.concat((mon_ds.cocco_P_lim_Cweight_avg_100m, mon_ds.cocco_Fe_lim_Cweight_avg_100m, mon_ds.cocco_N_lim_Cweight_avg_100m, mon_ds.cocco_C_lim_Cweight_avg_100m),dim='nutrient')
most_lim_sp=limarray_sp.argmin(dim='nutrient', skipna=False)
most_lim_diat=limarray_diat.argmin(dim='nutrient', skipna=False)
most_lim_diaz=limarray_diaz.argmin(dim='nutrient', skipna=False)
most_lim_cocco=limarray_cocco.argmin(dim='nutrient', skipna=False)
mask = np.isnan(ds.sp_N_lim_surf.squeeze())
fig = plt.figure(figsize=(12,23))
for row in np.arange(1,13):
ts=row-1
plot = row*3 - 2
#row 1 Jan
ax = fig.add_subplot(12,3,plot, projection=ccrs.Robinson(central_longitude=305.0))
ax.set_title('Diat biomass-weighted nut lim', fontsize=12)
lon, lat, field = utils.adjust_pop_grid(lons, lats, most_lim_diat.isel(month=ts).where(~mask))
pc=ax.pcolormesh(lon, lat, field, cmap=plt.cm.get_cmap('Dark2_r',4),vmin=-0.5,vmax=3.5,transform=ccrs.PlateCarree())
colorbar_specs = {'ticks' : np.arange(0,4,1)}
plot = row*3 - 1
ax = fig.add_subplot(12,3,plot, projection=ccrs.Robinson(central_longitude=305.0))
ax.set_title('SP biomass-weighted nut lim', fontsize=12)
lon, lat, field = utils.adjust_pop_grid(lons, lats, most_lim_sp.isel(month=ts).where(~mask))
pc=ax.pcolormesh(lon, lat, field, cmap=plt.cm.get_cmap('Dark2_r',4),vmin=-0.5,vmax=3.5,transform=ccrs.PlateCarree())
colorbar_specs = {'ticks' : np.arange(0,4,1)}
plot = row*3
ax = fig.add_subplot(12,3,plot, projection=ccrs.Robinson(central_longitude=305.0))
ax.set_title('Cocco biomass-weighted nut lim', fontsize=12)
lon, lat, field = utils.adjust_pop_grid(lons, lats, most_lim_cocco.isel(month=ts).where(~mask))
pc=ax.pcolormesh(lon, lat, field, cmap=plt.cm.get_cmap('Dark2_r',4),vmin=-0.5,vmax=3.5,transform=ccrs.PlateCarree())
colorbar_specs = {'ticks' : np.arange(0,4,1)}
fig.subplots_adjust(right=0.8)
cbar_ax = fig.add_axes([0.85, 0.15, 0.05, 0.7])
cbar = fig.colorbar(pc, cax=cbar_ax,**colorbar_specs)
cbar.ax.set_yticklabels(['P lim', 'Fe lim', 'N lim','SiO3/C lim']);